3.8. GENWIG: Generate wig data
The GENWIG mode generates wig data of ChIP/Input enrichment and p-value distributions. This mode is useful to analyze these distributions with Python and R by the user.
Here we use the yeast data used in PC_ENRICH: Enrichment visualization. To generate bigWig files of ChIP/Input enrichment for three sample pairs, type:
dir=parse2wigdir+
drompa+ GENWIG \
-i $dir/YST1019_Gal_60min.100.bw,$dir/YST1019_Gal_0min.100.bw,YST1019_Gal \
-i $dir/YST1019_Raf_60min.100.bw,$dir/YST1019_Raf_0min.100.bw,YST1019_Raf \
-i $dir/YST1053_Gal_60min.100.bw,$dir/YST1053_Gal_0min.100.bw,YST1053_Gal \
-o drompa-yeast --gt genometable.sacCer3.txt --outputformat 3 --outputvalue 0
then “drompa-yeast.YST1019_Gal.enrich.100.bw”, “drompa-yeast.YST1019_Raf.enrich.100.bw” and “drompa-yeast.YST1053_Gal.enrich.100.bw” are generated.
The detail of --outputvalue option:
--outputvalue 0 (default): ChIP/Input enrichment
--outputvalue 1: P-value (ChIP internal)
--outputvalue 2: P-value (ChIP/Input enrichment
and --outputformat option:
--outputformat 0: compresed wig (.wig.gz)
--outputformat 1: uncompresed wig (.wig)
--outputformat 2: bedGraph (.bedGraph)
--outputformat 3 (default): bigWig (.bw)
3.8.1. Tutorial
This is an example to generate a pdf file:
dir=parse2wigdir+
gene=data/S_cerevisiae/SGD_features.tab
gt=data/genometable/genometable.sacCer3.txt
drompa+ PC_ENRICH \
-i $dir/YST1019_Gal_60min.100.bw,$dir/YST1019_Gal_0min.100.bw,YST1019_Gal \
-i $dir/YST1019_Raf_60min.100.bw,$dir/YST1019_Raf_0min.100.bw,YST1019_Raf \
-i $dir/YST1053_Gal_60min.100.bw,$dir/YST1053_Gal_0min.100.bw,YST1053_Gal \
-o drompa-yeast --gt $gt -g $gene --gftype 2 --showpenrich 1 --showpinter 1 \
--scale_ratio 5 --ls 200 --sm 10 --lpp 3
To generate the bigWig files shown in this pdf file, use GENWIG command as below:
dir=parse2wigdir+
gt=../data/genometable/genometable.sacCer3.txt
drompa+ GENWIG \
-i $dir/YST1019_Gal_60min.100.bw,$dir/YST1019_Gal_0min.100.bw,YST1019_Gal \
-i $dir/YST1019_Raf_60min.100.bw,$dir/YST1019_Raf_0min.100.bw,YST1019_Raf \
-i $dir/YST1053_Gal_60min.100.bw,$dir/YST1053_Gal_0min.100.bw,YST1053_Gal \
-o drompa-yeast --gt $gt --outputformat 3 --outputvalue 0
drompa+ GENWIG \
-i $dir/YST1019_Gal_60min.100.bw,$dir/YST1019_Gal_0min.100.bw,YST1019_Gal \
-i $dir/YST1019_Raf_60min.100.bw,$dir/YST1019_Raf_0min.100.bw,YST1019_Raf \
-i $dir/YST1053_Gal_60min.100.bw,$dir/YST1053_Gal_0min.100.bw,YST1053_Gal \
-o drompa-yeast --gt $gt --outputformat 3 --outputvalue 1
drompa+ GENWIG \
-i $dir/YST1019_Gal_60min.100.bw,$dir/YST1019_Gal_0min.100.bw,YST1019_Gal \
-i $dir/YST1019_Raf_60min.100.bw,$dir/YST1019_Raf_0min.100.bw,YST1019_Raf \
-i $dir/YST1053_Gal_60min.100.bw,$dir/YST1053_Gal_0min.100.bw,YST1053_Gal \
-o drompa-yeast --gt $gt --outputformat 3 --outputvalue 2
Verify the generated bigWig files can generate the same distribution:
s=""
for sample in YST1019_Gal YST1019_Raf YST1053_Gal; do
for str in pinter penrich enrich; do
s="$s -i drompa-yeast.$sample.$str.100.bw"
done
done
drompa+ PC_SHARP $s -o drompa-yeast_genwig --gt $gt -g $gene --gftype 2 \
--scale_tag 5 --ls 200 --sm 10 --lpp 3
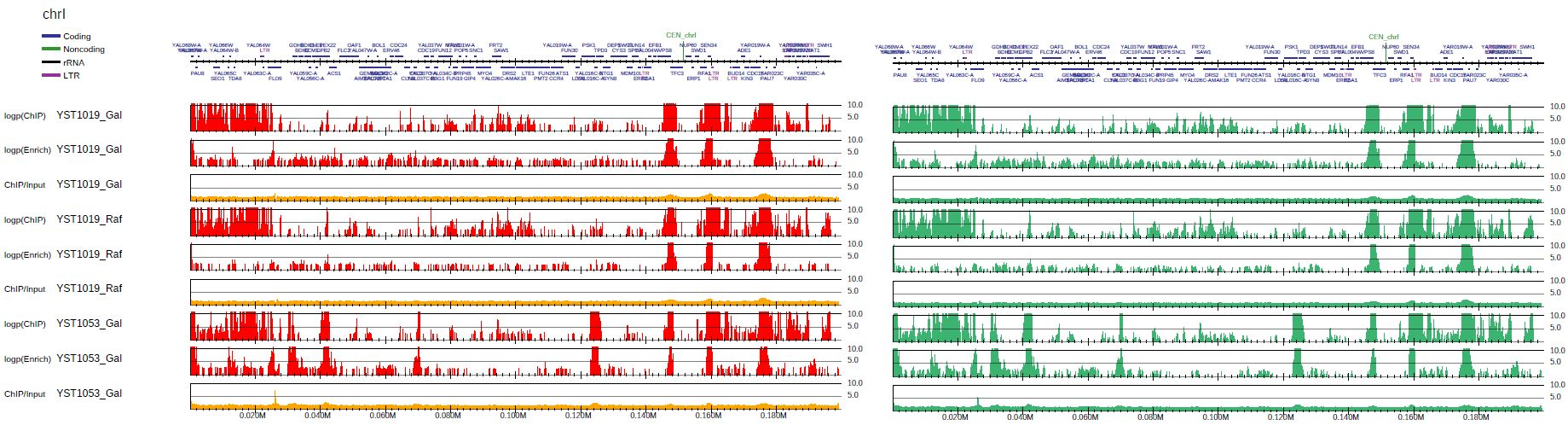
Fig. 3.29 The original pdf file (left) and the pdf using bigWig files generated by GENWIG command (right).