3.4. GV: chromosome-wide overview
A 100k-bp bin is recommended for the GV mode. Run parse2wig+ to make bigWig files for a 100k-bp bin:
$ parse2wig+ -i H3K4me3.bam -o H3K4me3 --gt genometable.txt -n GR --binsize 100000
$ parse2wig+ -i H3K27me3.bam -o H3K27me3 --gt genometable.txt -n GR --binsize 100000
$ parse2wig+ -i H3K36me3.bam -o H3K36me3 --gt genometable.txt -n GR --binsize 100000
$ parse2wig+ -i Input.bam -o Input --gt genometable.txt -n GR --binsize 100000
Then, run drompa+:
$ dir=parse2wigdir+
$ drompa+ GV \
-i $dir/H3K4me3.100000.bw,$dir/Input.100000.bw,H3K4me3 \
-i $dir/H3K27me3.100000.bw,$dir/Input.100000.bw,H3K27me3 \
-i $dir/H3K36me3.100000.bw,$dir/Input.100000.bw,H3K36me3 \
-o drompaGV --gt genometable.txt
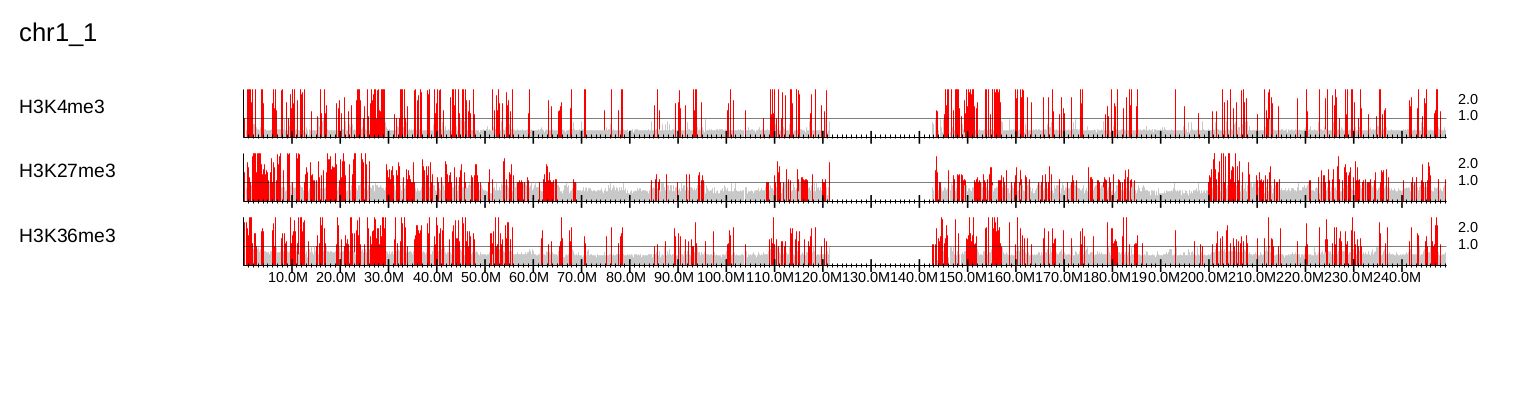
Fig. 3.20 Chromosome-wide overview.
The GV mode simply highlights in red the bins containing ChIP/Input enrichments above the middle of the y-axis (specified via the --scale_ratio).
3.4.1. Visualization with annotation
The GV mode can include various annotations::
$ dir=parse2wigdir+
$ drompa+ GV \
-i $dir/H3K4me3.100000.bw,$dir/Input.100000.bw,H3K4me3 \
-i $dir/H3K27me3.100000.bw,$dir/Input.100000.bw,H3K27me3 \
-i $dir/H3K36me3.100000.bw,$dir/Input.100000.bw,H3K36me3 \
-o drompaGV-K562_2 --gt genometable.txt \
--GC GCcontents --gcsize 500000 \ #
--GD genedensity --gdsize 500000 \ # Gene-density directory and window size (500 kbp)
--ideogram ../data/ideogram/hg19.tsv # ideogram
where options --GC -gcsize and --GD --gdsize specify the GC content directories and gene density with window size (500 kbp), respectively.
This command can work in the “tutorial” directory.
Ideogram data for several species are available in the “data/ideogram” directory.
The result is shown in Fig. 3.21.
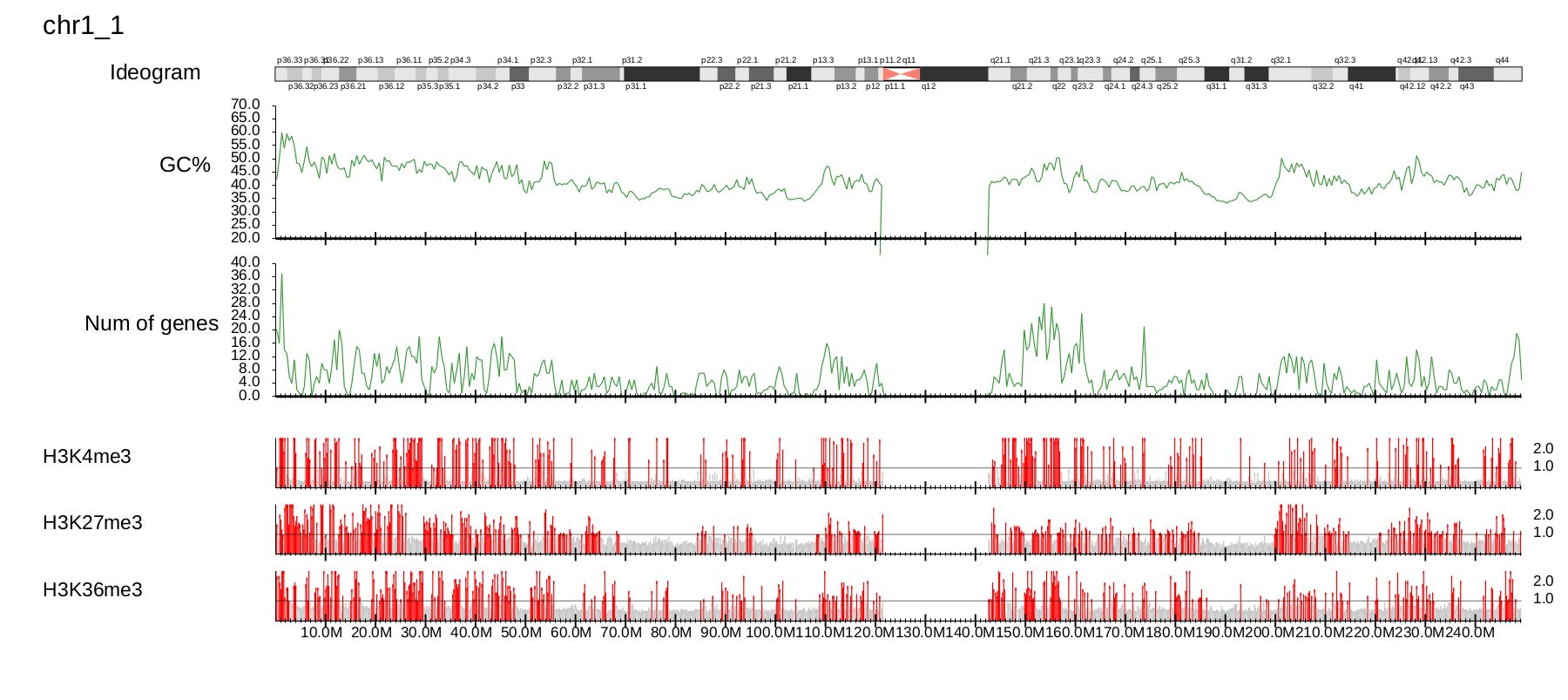
Fig. 3.21 Visualization with annotation.